Variable Alignment Settings
The Variable Alignment Settings interface allows modification of the Peak Alignment settings preprocessing method.
Peak Alignment adjusts data for shifting of the variable axes (usually when the variables are all from a continuous tuned source or detector and information "shifts" between variables as a result of mis-calibration or other instrumental or physical effects.
The interface allows selection between two alignment algorithms: Correlation Optimized Warping (COW) and Peak Alignment.
Correlation Optimized Warping
COW (Correlation Optimized Warping) - performs a piece-wise transformation of each sample adjusting the segments to best correlate to a target sample.
When selected, the user can select:
- Segment Length - size of the windows that the variables will be split into. Larger values allow for larger linear shifts (slack, see below, is limited to half the segment length, so larger segments allow larger shifts). Smaller values allow for more non-linear shifts (achieved by a piece-wise linear approximation) but limit the total possible shift and may cause some segments to lack sufficient information to correlate to.
- Slack - the maximum shift (in variables) which each segment can be shifted. Smaller values constrain the shift and reduce artifacts. Larger values allow for correcting for increasingly large shifts.
- Target Sample - defines which sample from the calibration set should be used as a target to correlate to. If Auto is chosen, the first calibration sample is used.
For more details, see Cow
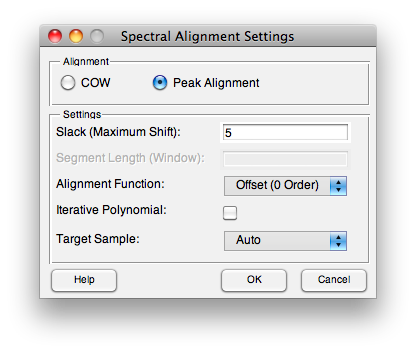
Peak Alignment
The Peak Alignment algorithm is based on the registerspec function. It locates peaks in both the target sample and the sample to be shifted (a "new sample"). These peak positions are then used to fit and apply a polynomial (of any order) or a piece-wise spline to align the new sample to the target sample.
When calibrating using this preprocessing, it first locates "acceptable peaks" in the target sample. If "Auto" is chosen for the target sample, then all calibration samples are used and only peaks that show insignificant shift (see "Slack" below) throughout the entire calibration set are used to align new samples. Otherwise, all the peaks in a specified target sample are used to align, as long as a corresponding peak is found in the new sample.
The settings include:
- Slack - Maximum amount of shift that is permitted before a peak in the new sample is considered "too shifted to use". When searching for the target peaks in "auto" target mode, this defines the maximum amount that a peak can shift before it is considered not appropriate to use for alignment. When aligning a new sample to the previously located peaks, this defines how far a given peak can be shifted before it is ignored during the polynomial alignment.
- Alignment Function - Either the order of polynomial to use in alignment of the located peaks or "Piece-wise Spline" to use a constrained pchip spline to align segments. The pchip spline is useful when there are many reference peaks and the axis may have non-linear shifts.
- Iterative Polynomial - When a polynomial Alignment Function is selected, enabling Iterative Polynomial will cause the alignment algorithm to repeat the alignment starting with a zero-order polynomial (shift) and increasing the order with each pass until the order specified in Alignment Function is reached. This method is slower but works better for badly shifted samples and/or higher-order polynomials.
- Target Sample - as described above, this defines what sample(s) the peak finding algorithm will search for "stable" peaks. If a single sample is selected, only that sample will be searched for peaks and all found peaks will be used to align new samples. If "Auto" is selected, only the peaks that are stable through all the samples will be used to align new samples.