Batchmaturity
Contents |
Purpose
Batch process model and monitoring, identifying outliers.
Synopsis
- model = batchmaturity(x,ncomp_pca,options);
- model = batchmaturity(x,y,ncomp_pca,options);
- model = batchmaturity(x,y,ncomp_pca,ncomp_reg,options);
- pred = batchmaturity(x,model,options);
- pred = batchmaturity(x,model);
Description
Analyzes multivariate batch process data to quantify the acceptable variability of the process variables during normal processing conditions as a function of the percent of batch completion. The resulting model can be used on new batch process data to identify measurements which indicate abnormal processing behavior (See the pred.inlimits field for this indicator.)
The progression through a batch is described in terms of the "Batch Maturity", BM, which is often defined in terms of percentage of completion of the process. In the BM analysis method a PCA model is built on the calibration batch dataset and the samples' PCA scores are binned according to the samples' BM values. The resulting model contains confidence limits on PCA scores as a function of batch maturity and reflect the normal range of variability of the data at any stage of progression through the batch. If one of the measured variable represents BM, or if BM is a known function of the measured variables, then a sample from a new batch can be tested against the model's score limits at its known BM value to see if it is normal sample or not. However, BM is often not measurable or known as a function of the measured variables in new batches. Instead, this relationship is estimated by building a PLS model using the calibration dataset where BM is provided. The PLS model is then used to predict the BM value for any sample.
For assistance in preparing batch data for use in Batchmaturity please see bspcgui.
Methodology
Given multivariate X data and a Y variable which represents the corresponding state of batch maturity (BM) build a model by:
- Build a PLS model on X and Y using specified preprocessing. Use its self-prediction of Y, ypred, as the indicator of BM.
- Simplify the X data by performing PCA analysis (with specified preprocessing). We now have PC scores and a measure of BM (ypred) for each sample.
- Sort the samples to be in order of increasing BM. Calculate a running-mean of each PC's ordered scores ("smoothed score means"). Calculate deviations of scores from the smoothed means for each PC.
- Form a set of equally-spaced BM values over the range (BMstart, BMend). For each BM point, find the n samples which have BM closest to that value.
- For each BM point, calculate low and high score limit values corresponding to the cl/2 and 1-cl/2 percentiles of the n sample score deviations just selected (repeat for each PC). Add the smoothed scores to these limits to get the actual limits for each PC at each BM point. These BM points and corresponding low/high score limits constitute a lookup table for score limits for each PC in terms of BM value.
- The score limits lookup table contains upper and lower score limits for each PC, for every equally-spaced BM point over the BM range.
- The batch maturity model contains the PLS and PCA sub-models and the score limits lookup table. It is applied to a new batch processing dataset, X1, by applying the PLS sub-model to get BM (ypred), then applying the PCA sub-model to get scores. The upper and lower score limits (for each PC) for each sample are obtained by using the sample's BM value and querying the score limits lookup table. A sample is considered to be an inlier if its score values are within the score limits for each PC.
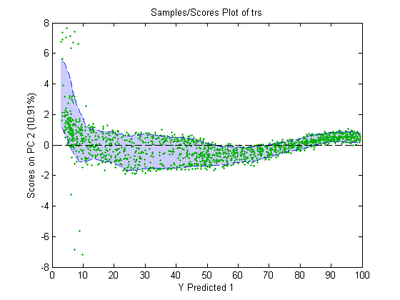
Fig. 1 shows an example of the Batch Maturity Scores Plot (obtained from the BATCHMATURITY Analysis window's Scores Plot). This shows the second PC scores upper and lower limit as a function of BM as calculated form the "Dupont_BSPC" demo dataset using cl = 0.9 and step 2 only from batches 1 to 36. These batches had normal processing conditions so the shaded zone enclosed by the limit lines indicates the range where a measured sample's PC=2 scores should occur if processing is evolving "normally". Similar plots result for the other modeled PCs. The data points shown are the PCA model scores, which are accessible from the batchmaturity model or pred's t field.
- Fig. 1. Batchmaturity Scores Plot.
Comments
An anomalous sample will be identified as such in a batch maturity analysis except in the rare case where the anomaly happens to look like a normal sample from a different stage of the process evolution. This should be a rare occurrence in a batch process dataset especially when there are more than one or two variables in the dataset.
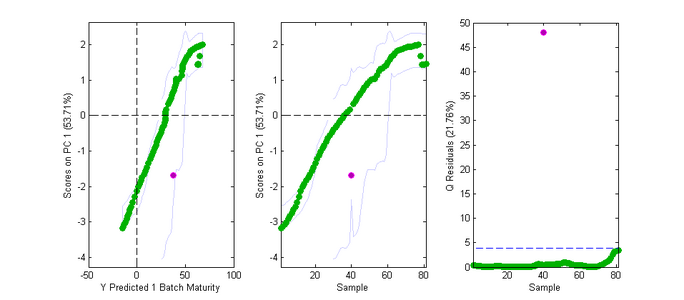
Fig. 2 shows batch maturity results for a dataset having 81 samples with 5 variables. Sample #40 is an anomaly as is shown in plots of PC 1 vs. pred BM (Fig. 2a,b), or a plot of the PCA Q residuals versus sample number (shown) or pred BM (not shown). The bad sample is not well represented by the PCA model so its Q is large.
- Fig. 2. Batchmaturity Scores Plot.
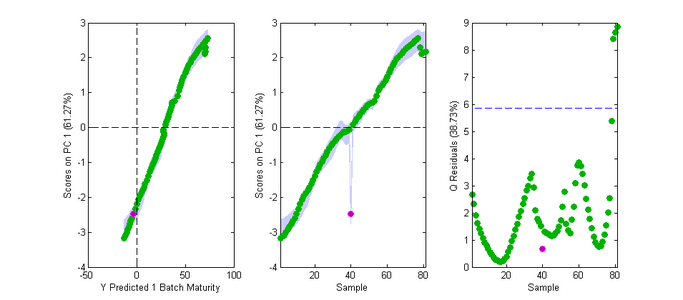
Fig. 3 shows similar results but where sample #40's data were replaced by sample #10's, making sample #40 an anomaly for that stage of the process. This is not shown by the batch maturity analysis, however, because the predicted BM value from the PLS model for sample #40 is the same as for sample #10, so it is treated as another sample identical to sample #10. Thus it does not show up as unusual in Fig. 3a, nor in Fig. 3c because its Q is not unusual compared to all the other samples. It does appear unusual in Fig. 3b where the temporal order of samples is used as the x-axis - however Batch Maturity models do not take this temporal information into account and, as such, there is usually no way to use a BM model to detect such simple "misplacement" anomalies.
- Fig. 3. Batchmaturity Scores Plot.
Inputs
- x = X-block (2-way array class "double" or "dataset").
- y = Y-block (vector class "double" or "dataset").
- ncomp_pca = Number of components to to be calculated in PCA model (positive integer scalar).
- ncomp_reg = Number of latent variables for regression method.
Outputs
- model = standard model structure containing the PCA and Regression model (See MODELSTRUCT).
- pred = prediction structure contains the scores from PCA model for the input test data as pred.t.
Model and pred contain the following fields which relate to score limits and whether samples are within normal ranges or not:
- limits : struct with fields:
- cl: value used for cl option
- bm: (1 x bmlookuppts) bm values for score limits
- low: (nPC x bmlookuppts) lower score limit of inliers
- high: (nPC x bmlookuppts) upper score limit of inliers
- median: (nPC x bmlookuppts) median trace of scores
- inlimits : (nsample x nPC) logical indicating if samples are inliers.
- t : (nsample x nPC) scores from the PCA submodel.
- t_reduced : (nsample x nPC) scores scaled by limits, with limits -> +/- 1 at upper/lower limit, -> 0 at the median score.
- submodelreg : regression model built to predict bm. Only PLS currently.
- submodelpca : PCA model used to calculate X-block scores.
Options
options = a structure array with the following fields:
- regression_method : [ {'pls'} ] A string indicating type of regression method to use. Currently, only 'pls' is supported.
- preprocessing : { [] } preprocessing structure goes to both PCA and PLS. PLS Y-block preprocessing will always be autoscale.
- zerooffsety : [ 0 | {1}] transform y resetting to zero per batch
- stretchy : [ 0 | {1}] transform y to have range=100 per batch
- cl : [ 0.90 ] Confidence limit (2-sided) for moving limits (defined as 1 - Expected fraction of outliers.)
- nearestpts : [{25}] number nearby scores used in getting limits
- smoothing : [{0.05}] smoothing of limit lines. Width of window used in Savgol smoothing as a fraction of BM range.
- bmlookuppts : [{1001}] number of equally-spaced points in BM lookup table mentioned in Methodology Step 4 above. Default gives lookup values spaced every 0.1% over the BM range.
- plots : [ 'none' | 'detailed' | {'final'} ] governs production of plots when model is built. 'final' shows standard scores and loadings plots. 'detailed' gives individual scores plots with limits for all PCs.
- waitbar : [ 'off' | {'auto'} ] governs display of waitbar when calculating confidence limits ('auto' shows waitbar only when the calculation will take longer than 15 seconds)